Protein Aggregation Pathway Selection
Neurodegeneration is associated with a class of diseases known as foldopathies, in which specific proteins (e.g. α-synuclein, huntington, amyloid-β or tau) misfold, oligomerize and aggregate to form fibrils. Of these, the Han lab has focused mostly on tau. The mechanism of tau toxicity is still unclear, but it is presumed that pathological tau “prions” within neurons are capable of seeding functional tau in neighboring neurons into fibrillar strains resembling the pathological property of the initial tau “prion”, and that this process is an obligatory component of tau-mediated pathogenesis. However, neither the identity of the tau “prion” seed, nor the molecular basis of the seeding process is known. Also, structural convergence to a homogeneous tau fibril structure has never been achieved by seeding, let alone to Alzheimer Disease (AD), Pick's Disease (PiD) or Cerebral Traumatic Encephalopathy (CTE) disease phenotypic tau fibril structures, recently solved by cryo-EM. The Han lab focuses on solving the fundamental mechanism to select specific tau conformers, to achieve templating, and to ultimately populate distinct tau fibrils through seeded aggregation. En route to these goals, the Han lab studied tau peptides (12-, 14-, 43-, 187-mer) and full-length tau (2N4R and 3R), and learned that deliberate changes to the conformation and population of tau monomers in solution state, and in the early stages of aggregation, affect their fibril formation propensity downstream. These findings suggest that therapeutic intervention may focus on disrupting the earliest aggregation events occurring in solution. We found that heparin achieves conformational templating, but is an imperfect template and dominant cofactor that generates heterogeneous fibrils. RNA cofactors and liquid-liquid phase separation conditions were found to lower the seeding barrier, while well-defined fibril seeds can achieve partial structural convergence, but only in the presence of certain cofactors. Our goal is to understand common biophysical features and conformational signatures of a tau seed that can nucleate templated aggregation, and to ultimately achieve the selection of disease phenotypic protein aggregation pathways.
Protein Surface Engineering
The emergence of high-resolution structures of biological building blocks is revolutionizing the way we can study the protein machinery. Resolving the structure of proteins, protein aggregates and molecular complexes represents a necessary starting point to uncover their activity and function. Yet this knowledge offers no direct predictive insights into how proteins reach their structure and achieve their function. The key knowledge gap lies in the translation of protein structure and surface chemistry into protein property and function. This knowledge gap spans a broad scope in molecular biochemistry.
For example, a property as fundamental as protein stability or solubility cannot be a priori predicted from a known protein structure with our current understanding and tools. This is underscored by observations that single mutations on the surface of proteins with no or minimal structural changes can dramatically alter the solubility or aggregation propensity of a protein. Another example is the aggregation of intrinsically disordered proteins (IDPs) that lack a well-defined 3D structure and are involved in many neurodegenerative diseases. The stability, or for that matter aggregation propensity, of IDPs is even harder to predict than of folded proteins. There are many familial mutations, often defined by a single amino acid mutation that are known to dramatically alter the aggregation propensity. Yet, the molecular basis of these effects, e.g. whether the mutation alters the conformational equilibrium or protein hydration landscape, is not understood. Our hypothesis is that mutations alter the free energy of specific conformers through multiple mechanisms including the modification of their solvation free energy.
Our approach is to uncover the code for translating a range of protein surface structural properties into protein interaction and function. The Han lab is pursuing this translation using biochemical, molecular biological, and state-of-the-art characterization tools that capture surface structural properties with high information content and resolution, including of protein surface-coupled water, protein dynamics, and protein conformation. A focus lies on rationalizing the relation between solvation and protein function. Several of these studies are made possible by the development of novel spectroscopic tools by the Han lab for detecting equilibrium hydration water dynamics and topology, and weak protein interactions by Overhauser dynamic nuclear polarization (ODNP)-amplified nuclear magnetic resonance (NMR) relaxometry. The Han lab also relies on complementary techniques to map out structure, dynamics and interaction by paramagnetic resonance (EPR), pulsed double electron-electron resonance (DEER) and solid-state MAS NMR with dynamic nuclear polarization (DNP) enhancement. The Han lab's broader vision to uncover the design rules for interaction and assembly of nature’s molecular machines.
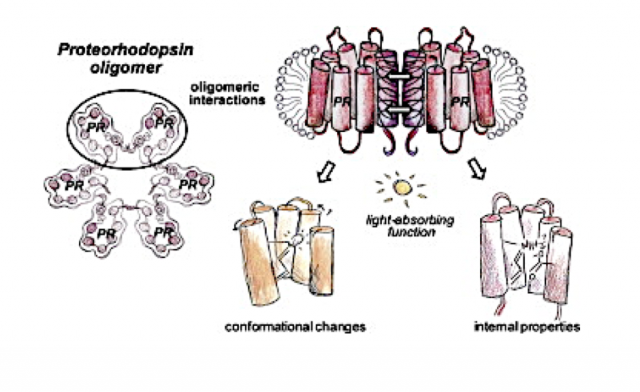
Membrane Protein Oligomer Structure-Function
The plasma membrane is the crucial interface between the cell and its exterior, packed with embedded proteins experiencing simultaneous protein-protein and protein-membrane interactions. A prominent example of cell membrane complexity is the assembly of transmembrane proteins into oligomeric structures, with potential functional consequences that are not well understood. From the study of proteorhodopsin (PR), we find evidence that the inter-protein interaction modulated by self association yields functional changes observable from the protein interior. We also demonstrate that the oligomer is a physiologically relevant form of PR, as crosslinking of recombinantly expressed PR reveals an oligomeric population within the E. coli membrane (putatively hexameric).
Equipped with ODNP methods that exploit surface water structure to elucidate protein interfaces and secondary protein structure and membrane insertion, and pulsed DEER and CW EPR tools to measuring 1-7 nm distances, we mapped out the structural transformation of PR upon light activation, the oligomeric structure of PR in various detergent complexes and in liposome environments, and the functional consequence of PR oligomerization in recent studies. These methods are extended to study the oligomer structure-function relationship of the G-protein coupled receptor (GPCRs), adenosine A2A receptors, in collaboration with O’Malley. GPCR oligomerization is thought to broaden the range of cell signaling, but their study requires novel characterization tools and concepts. We found that specific segments of the intrinsically disordered C-terminus of A2A is responsible for A2A dimer formation, while all existing structure studies of A2A are on the C-terminal truncated variant. The structure-function study of the A2A dimer is a prime goal. With these studies, the Han lab is testing a potentially general design principle for transmembrane protein function tuning by specific oligomeric contacts modulated by protein expression or membrane composition.

Biopolymer Liquid-Liquid Phase Separation
The Han lab has long-standing interest in the liquid-liquid phase separation (LLPS) of proteins and polyelectrolytes, which is a fascinating phenomenon long known to the biopolymer community, and now is drawing significant attention in the field of neurodegeneration as a potential regulatory mechanism for protein segregation, aggregation and dissolution. The Han lab's quantitative studies of LLPS enabled by advanced characterization methods that were initially focused on mussel foot proteins and polymers helped us gain a strong background and intuition about this phenomenon. Recent studies of the Han lab established that LLPS of tau-RNA association is an equilibrium state with an easily accessible phase boundary by physiological factors, including temperature, salt and tau concentrations, and is entropy-driven. Guided by our phase diagram, we show that tau can be driven toward LLPS under live cell coculturing conditions with rationally chosen experimental parameters. The Han lab's current focus is to unravel the role of LLPS of tau, either by complex coacervation or by simple coacervation, in protecting, modulating or facilitating tau aggregation towards strain-specific amyloid fibrils.